Antibiotics are minor miracles. They actually prevent people from dying of disease (most drugs are treatments rather than cures), quite possibly saving more lives than any other product of human innovation.
During the ‘golden age’ of antibiotic discovery in the 1940s to 1970s, scientists discovered hundreds, if not thousands, of new antibiotics, mainly natural products isolated from microbiological sources. The best of these were developed into drugs able to cure life-threatening infections. These early discoveries involved collaborative research largely unencumbered by confidential disclosure and material transfer agreements, with academic and industrial researchers working together to help save lives.
Unfortunately, bacteria have rapidly adapted to every new antibiotic introduced into clinics over the past 70 years. This has led to an ‘arms race’ in which scientists have continually developed more potent or novel antibiotics to stay a step ahead of the growing bacterial resistance. However, our discoveries of new antibiotics have not kept up with bacteria’s ability to become resistant to existing therapies. A growing number of infections, particularly in countries such as Greece and India, where antibiotic misuse is rampant, are proving fatal because they are unable to be successfully treated by any antibiotic. These extremely drug-resistant bacteria are becoming increasingly common, readily swapping their resistance genes with other bacteria, and being carried around the world by international travellers. If this continues, infections that were previously innocuous may lead to death. If we are unable to use antibiotics preventatively, most surgeries will be impossible.
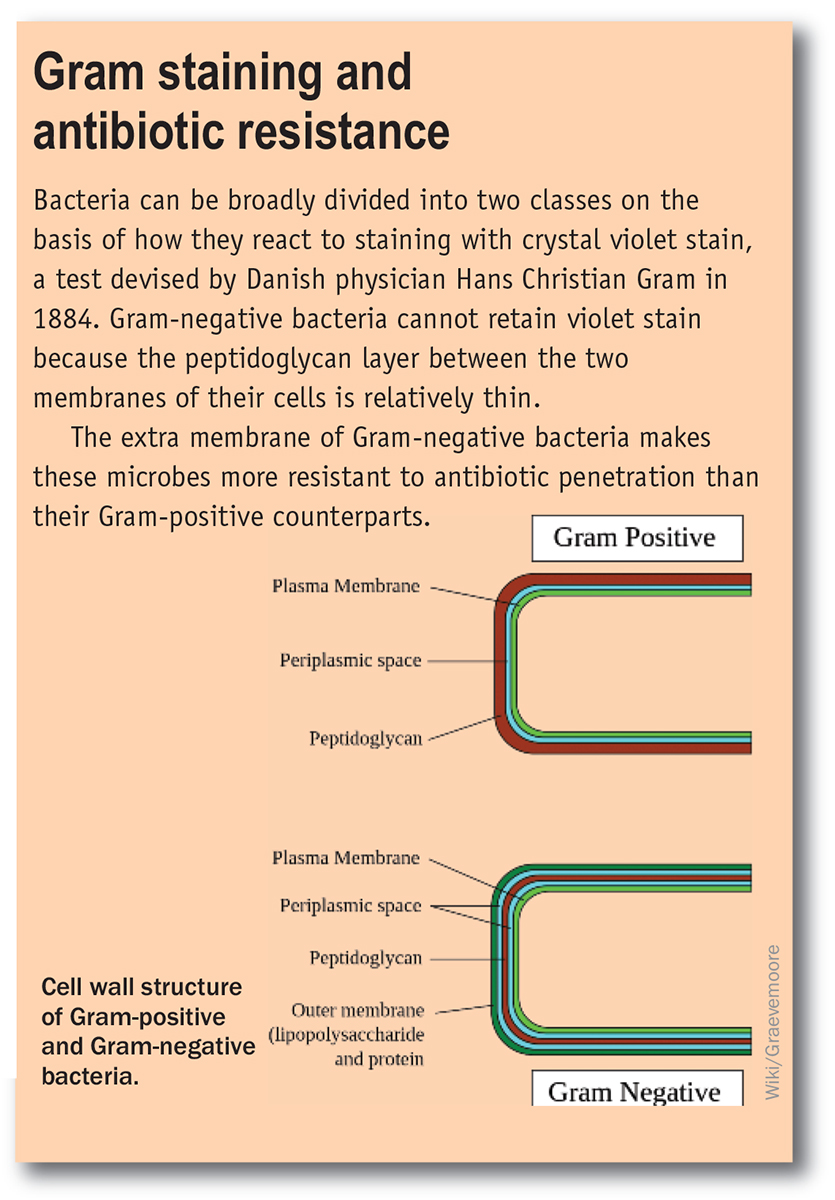
How has this happened? One cause is the misuse and overuse of antibiotics. Antibiotics have been widely used as growth promoters in animal feed. They are often prescribed to treat viral infections, where they have no effect. Patients often do not complete a full course of antibiotics, allowing a resistant subpopulation to survive. Poor hospital management, with a lack of handwashing and other appropriate infection controls, provides localised centres where resistant species can become established and thrive. The other main issue is a lack of new antibiotics in the clinical pipeline, particularly for the more difficult-to-treat Gram-negative infections (see box below). The traditional source of new antibiotics, the large pharmaceutical companies, have largely abandoned antimicrobial research because research in this field is hard to justify economically – they can sell a new anticancer therapy that prolongs a life for nearly $US100 000 per year, but are unable to charge much more than $US5000 for a two-week course of antibiotics that can save a life.
There is a public misperception that we may not need antibiotics. Indeed, alternative approaches for treating infections do exist. Vaccines have proved remarkably effective at reducing infections for certain pathogens, such as the meningococcal vaccine preventing meningitis caused by Neisseria meningitides. Unfortunately, vaccines are highly specific for the bacteria they are designed for, and are preventative, not a treatment once infection has happened. Phage therapy, used for decades in Eastern European countries, is a potential approach enjoying a renaissance, but once again it is highly specific for individual pathogens: a phage designed to treat an E. coli infection will not work for an MRSA (methicillin-resistant Staphylococcus aureus) infection. ‘Natural’ therapies that kill bacteria, such as a thousand-year-old Medieval Anglo-Saxon onion and garlic eye remedy that was recently shown to kill MRSA, are sometimes covered in the popular press. Although many of these natural extracts can kill bacteria exposed on a surface, if they were used to treat an infection within the body, where life-threatening infections occur, they would either be inactivated by and/or be highly toxic to the body itself.
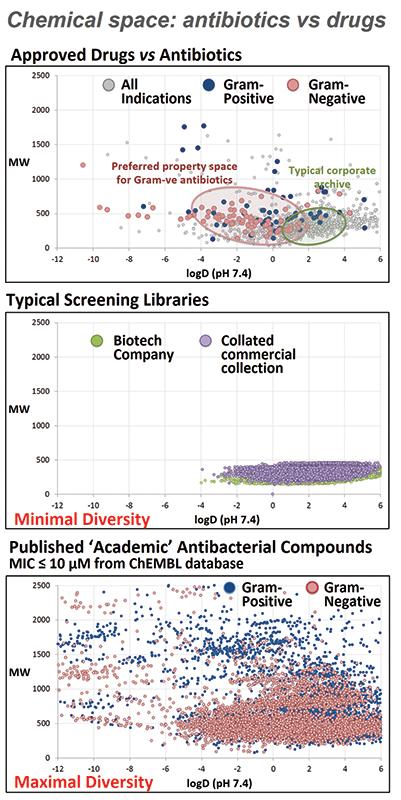
We need new ways to discover new antibiotics. Antibiotics are different from many other types of drugs, and a lot of the rules developed over the last 20 years for drug discovery projects that focus on ‘drug-like’ properties mean that potential antibiotics might be discarded before they are even tested. If you look at two common descriptors for ‘drug-like’ properties – size (molecular weight) and lipophilicity (log P) – most large pharmaceutical companies and commercially available screening libraries have a very limited distribution (see middle graph), as this range covers the chemical space of most orally available approved drugs (top graph). Approved antibiotics exhibit a much more diverse range (top graph), partly because many are administered intravenously. Published academic compounds that have reported antimicrobial activity show a similar diverse distribution (bottom graph), so if only those with ‘drug-like’ properties are considered, potential life-saving antibiotics may be missed.
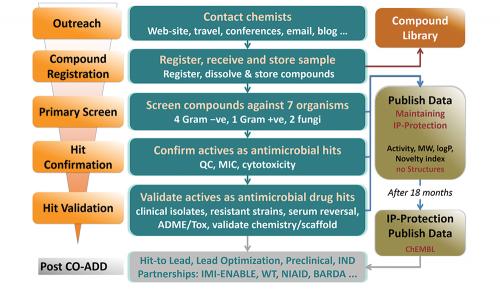
The Australian-based Community for Open Antimicrobial Drug Discovery (CO-ADD) is our attempt to develop a collaborative pipeline of new antibiotic candidates by mining the diverse chemical space of synthetic chemists around the world. Chemists make new molecules for all sorts of reasons, and many of these molecules have unusual structures. However, once made, many compounds are either thrown away or just get stored on shelves and in fridges in the backs of labs, and are certainly never tested for antimicrobial activity. With funding from the Wellcome Trust and support from the University of Queensland,
CO-ADD is offering free testing to see if compounds can kill any one of five key pathogenic bacteria or two fungi. Importantly, whoever submits the compound keeps all the rights to publish or patent, and develop any promising compounds. CO-ADD will use the screening data to generate a publically accessible database to allow other scientists to see what types of molecules have antimicrobial activity and, just as importantly, what types don’t.
Just 1 mg of compound is required for testing, or 50 µL of a 10 mM solution. Once received, compounds undergo an initial single concentration screen at 32 µg/mL to test their ability to inhibit the growth of any of five bacteria (one Gram-positive, MRSA, and four Gram-negative, Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa) or two fungi (Candida albicans, Cryptococcus neoformans) (see flowchart). Any ‘hits’ undergo a ‘hit confirmation’ assay, with dose ranging measurement of the minimum inhibitory concentration against the pathogens, counter-screening for cytotoxicity against mammalian cells, and quality control to confirm identity and purity. Compounds that pass these criteria are progressed to a more comprehensive evaluation in the ‘hit validation’ stage. All the data is returned to the submitting scientist, for them to publish, patent or develop.
CO-ADD has had a great reception from the scientific community, with more than 80 participating groups from 26 countries sending nearly 20 000 compounds in our first eight months, and over 300 000 additional compounds promised. We have run a ‘Thinkable’ competition (http://thinkable.org/competition/30) to award novel ideas for submissions. It’s still too early to see whether we’ll discover the next antibiotic, but we’ve already identified nearly 500 compounds with promising activity, which require further testing.
To foster the collaborative effort, CO-ADD has established contacts with a range of other organisations, including Compounds Australia, IMI-ENABLE (the European Union Innovative Medicines Initiative – European Gram-negative Antibacterial Engine), ChEMBL, ANTRUK (Antibiotic Resistance UK), NPS MedicineWise, the RACI, Royal Society for Chemistry and American Chemical Society. Notably, the output of CO-ADD (validated antimicrobial activity under standardised conditions, coupled with additional characterisation) is exactly the data package that IMI-ENABLE is asking for in submissions for hit-to-IND (investigational new drug) development. Looking to the future, CO-ADD is actively seeking additional funding to expand screening of this unique collection of compounds to include other neglected diseases such as tuberculosis, malaria, dengue and parasitic diseases such as leishmaniasis and Chagas disease.
CO-ADD relies on your help to be successful, so please dig into those dusty boxes of vials and send us your compounds! Students, set aside a sample of every compound you make, and send them in every few months. You could quite easily gain an additional publication for relatively little effort and, more importantly, contribute towards a solution that will save lives.